Proteins are the activated form of genetic information. There are approximately 20,000 different types of proteins in the human body. Proteins play many roles such as defense, transport, enzymes and communication.
The shape determines the functions of proteins. The shape of the proteins determines the way they are folded. The amino acid sequence and type involved determine the folding patterns. A mutation in DNA indirectly alters the function of the protein to be formed.
Since only 1 amino acid is changed in sickle cell anemia patients, hemoglobin protein folds differently.
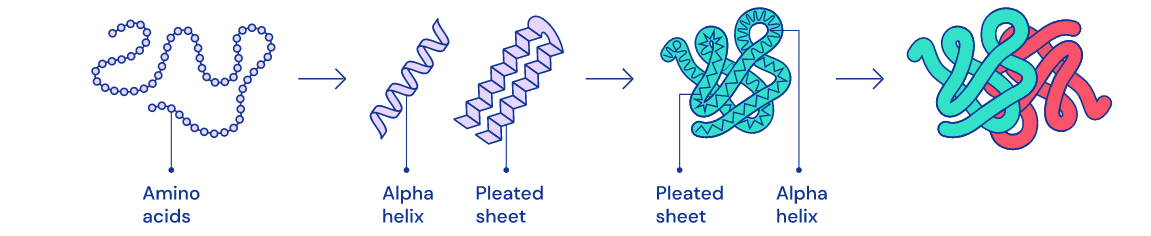
4 Stages Of Folding

Thanks to the ribosome, 20 different kinds of amino acids form proteins by arranging them end-to-end like a chain, and DNA has predetermined which amino acids will be sequenced in which order for each protein.
Folding basically takes place in 4 steps:
- In the first step, the protein is a straight chain of amino acids.
- In the second step the chain creates alpha helixes and beta layer structures
- In other steps, the protein becomes more and more organized. It can be thought of as creating words from letters, sentences and paragraphs from words.
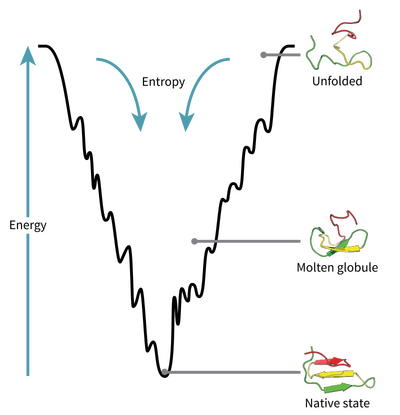
Levinthal’s paradox

A molecular biologist named Cyrus Levinthal calculated that it is near impossible (1/3¹⁹⁸ for a chain of 100 amino acids) for a protein to randomly fold into its correct shape.
For this reason, proteins must follow some rules when folding. Cell drawings made today are optimistic, in reality the inside of a cell is very complex and billions of collisions take place per second. Even though proteins are affected by these collisions as they fold, the forces in the optimization process are greater.
The primary chain can be thought of as different magnets attached end-to-end, when this chain is released the magnets will apply different forces to each other and eventually reach the lowest energy level.
Proteins try to reach the lowest energy level by minimizing the surface area, just like a water droplet being a sphere.
Teaching Biology to Machines
Competing with the name Alpha Fold, DeepMind won the CASP competition with 25 predictions, outperforming its closest competitor that could only guess 3 of the 43 proteins.Despite the lack of biochemistry experience on the DeepMind team,they defeated Giants like Novartis.
Proteins can be encoded according to the amino acids they contain, such as “VWDALRNETVKQR”. Then, with the coded information, specially designed artificial neural networks can be fed.
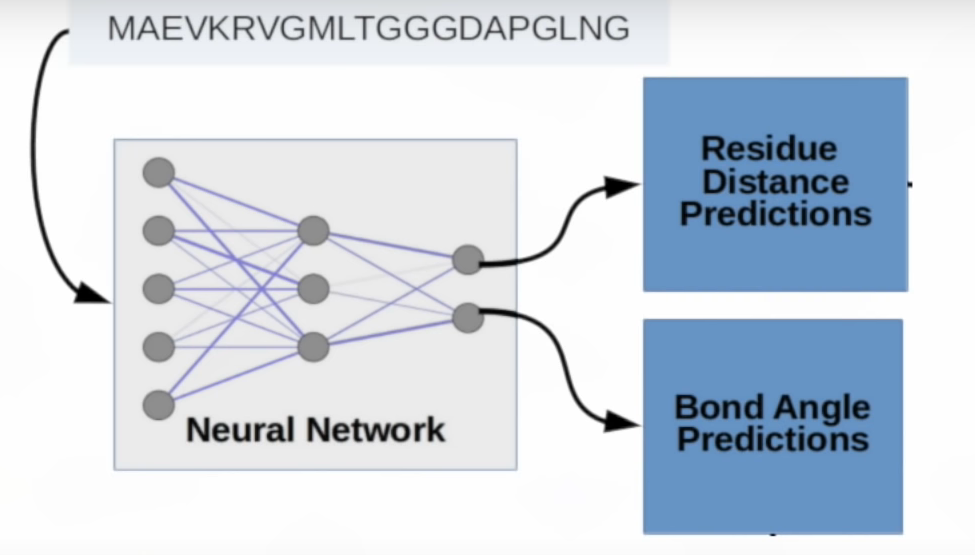
Two pieces of information are sufficient to predict the 3D shape of the protein:
- Distance between amino acids.
- Angle between chemical bonds. While 1 angle value is sufficient for 2 dimensions, 2 angle values are required in the 3rd dimension.
After the estimated distance and angle information, it can be visualized into proteins with special software.

In this process, the researchers developed special activation (Elu) functions and convolution (dilated) layers. Similar to biological processes, the structure of the molecule is determined by minimizing the potential energy by Gradient Descent.
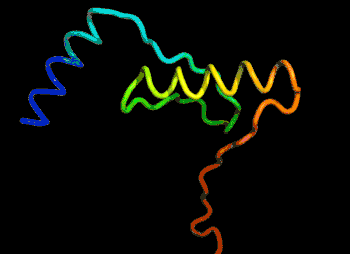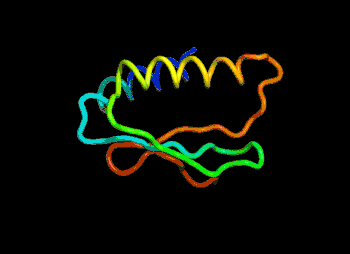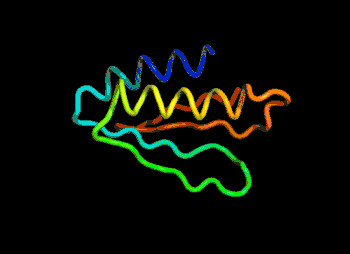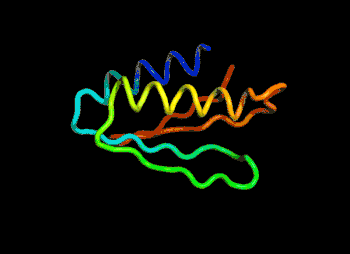
The most efficient technique that can be applied would be to find and modify the known protein with the closest sequence to the unknown protein in 3D shape. For example, if the unknown protein is “ACKLPWTSRQN”, the closest known is “ACKLPWTSRTQ”, successful predictions can be made by arranging the last 2 amino acid linkages.
Scope of application
- Special proteins can be developed that target viruses and mutant proteins.
- Alzheimer’s, Parkinson’s, Huntington’s disease and Cancer strains are thought to be linked to protein folding. Improving understanding of protein folding will accelerate future treatments.
— Sources —
2-Deepmind
Author

Ömer Özgür
AI TEAM